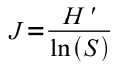DESCRIPTION
r.li.pielou calculates the "Pielou's diversity index" as:
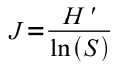
with:
- J: Pielou eveness index
- H': Shannon index
- S: number of classes
NOTES
Do not use absolute path names for the config and output
file/map parameters.
If the "moving window" method was selected in r.li.setup, then the
output will be a raster map, otherwise an ASCII file will be generated in
the folder C:\Users\userxy\.r.li\output\
(MS-Windows) or $HOME/.r.li/output/ (GNU/Linux).
If the input raster contains NULL value cells, r.li.pielou
returns -1 for these cells.
If you want to change these -1 values to NULL, run subsequently on the resulting map:
r.null setnull=-1 map=my_map
EXAMPLES
To calculate Pielou's diversity index on map my_map, using
my_conf configuration file (previously defined with
r.li.setup) and saving results in my_out, run:
r.li.pielou map=my_map conf=my_conf output=my_out
Forest map (Spearfish sample dataset) example:
g.region rast=landcover.30m -p
r.mapcalc "forests = if(landcover.30m >= 41 && landcover.30m <= 43,1,null())"
r.li.pielou map=forests conf=movwindow7 out=forests_pielou_mov7
r.univar forests_pielou_mov7
Forest map (North Carolina sample dataset) example:
g.region rast=landclass96 -p
r.mapcalc "forests = if(landclass96 == 5, 1, null() )"
r.li.pielou map=forests conf=movwindow7 out=forests_pielou_mov7
# verify
r.univar forests_pielou_mov7
r.to.vect input=forests output=forests feature=area
d.mon x0
d.rast forests_pielou_mov7
d.vect forests type=boundary
SEE ALSO
r.li - package overview
r.li.setup
REFERENCES
McGarigal, K., and B. J. Marks. 1995. FRAGSTATS: spatial pattern
analysis program for quantifying landscape structure. USDA For. Serv.
Gen. Tech. Rep. PNW-351. (PDF)
AUTHORS
Luca Delucchi and Duccio Rocchini, Fondazione E. Mach (Italy), based on the r.li.shannon code
developed by Serena Pallecchi student of Computer Science University of Pisa (Italy).
Last changed: $Date$